Week 5: Working with Tabular Data in Python with Pandas¶
This week, you will learn how to work with tabular data in Python.
Pandas is a an open source library providing high-performance, easy-to-use data structures and data analysis tools. Pandas is particularly suited to the analysis of tabular data, i.e. data that can can go into a table. In other words, if you can imagine the data in an Excel spreadsheet, then Pandas is the tool for the job.
A recent analysis of questions from Stack Overflow showed that python is the fastest growing and most widely used programming language in the world (in developed countries).
A follow-up analysis showed that this growth is driven by the data science packages such as numpy, matplotlib, and especially pandas.
The exponential growth of pandas is due to the fact that it just works. It saves you time and helps you do science more efficiently and effictively.
Pandas capabilities (from the Pandas website):¶
- A fast and efficient DataFrame object for data manipulation with integrated indexing;
- Tools for reading and writing data between in-memory data structures and different formats: CSV and text files, Microsoft Excel, SQL databases, and the fast HDF5 format;
- Intelligent data alignment and integrated handling of missing data: gain automatic label-based alignment in computations and easily manipulate messy data into an orderly form;
- Flexible reshaping and pivoting of data sets;
- Intelligent label-based slicing, fancy indexing, and subsetting of large data sets;
- Columns can be inserted and deleted from data structures for size mutability;
- Aggregating or transforming data with a powerful group by engine allowing split-apply-combine operations on data sets;
- High performance merging and joining of data sets;
- Hierarchical axis indexing provides an intuitive way of working with high-dimensional data in a lower-dimensional data structure;
- Time series-functionality: date range generation and frequency conversion, moving window statistics, moving window linear regressions, date shifting and lagging. Even create domain-specific time offsets and join time series without losing data;
- Highly optimized for performance, with critical code paths written in Cython or C.
- Python with pandas is in use in a wide variety of academic and commercial domains, including Finance, Neuroscience, Economics, Statistics, Advertising, Web Analytics, and more.
Grading:¶
- Total: 100
- Complete the example code: 30
- Exercise: 32
- Assignment: 28
- Comments clearly and concisely explain the purpose behind code: 10
Let's start by importing pandas library.
import pandas as pd
import numpy as np
1. Basic Pandas¶
1.1 Basic data structures in Pandas:¶
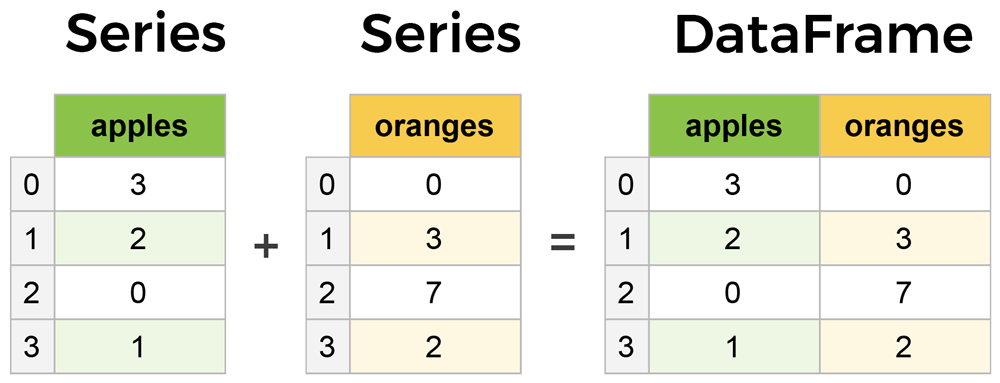
Pandas provides two types of classes for handling data:
- Series: a one-dimensional labeled array holding data of any type.
- DataFrame: a two-dimensional data structure that holds data like a two-dimension array or a table with rows and columns.
You can think of Pandas Dataframe as an Excel spreadsheet, and Series as one column of the the spreadsheet. Multiple series can be combined as a DataFrame.
from IPython.display import Image
Image(url='https://storage.googleapis.com/lds-media/images/series-and-dataframe.width-1200.png')
1.2 Pandas Series¶
Creating a Series by passing a list of values.
s = pd.Series([1, 3, 5, np.nan, 6, 8])
s
0 1.0 1 3.0 2 5.0 3 NaN 4 6.0 5 8.0 dtype: float64
By default, Pandas creates a default RangeIndex starting from 0.
Creating a Series by passing values and indices.
s = pd.Series([1, 3, 5, np.nan, 6, 8], index = ['A','B','C','D','E','F'])
s
A 1.0 B 3.0 C 5.0 D NaN E 6.0 F 8.0 dtype: float64
Pandas has plotting functions that can easily visualize the data.
s.plot(kind = 'bar')
<Axes: >
Arithmetic operations and most numpy function can be applied to Series.
An important point is that the Series keep their index during such operations.
np.sqrt(s)
A 1.000000 B 1.732051 C 2.236068 D NaN E 2.449490 F 2.828427 dtype: float64
We can also access the underlying index object if we need to:
s.index
Index(['A', 'B', 'C', 'D', 'E', 'F'], dtype='object')
Exercise 1: Create a Pandas series with values ranging from 0 to 6, and label the index as the day of week, starting from 'Sunday' to 'Saturday'. (3 points)¶
1.3 Indexing¶
We can get values back out using the index via the .loc attribute
s.loc['A']
np.float64(1.0)
Or by raw position using .iloc:
s.iloc[0]
np.float64(1.0)
We can pass a list or array to loc to get multiple rows back:
s.loc[['A','C']]
A 1.0 C 5.0 dtype: float64
We can also use the slice notation:
s.loc['A':'C']
A 1.0 B 3.0 C 5.0 dtype: float64
Exercise 2: Print out the last two elements of Series s (3 points)¶
hint: using slice notation and iloc
1.4 Pandas data structure: DataFrame¶
A more useful Pandas data structure is the DataFrame. A DataFrame is basically a bunch of series that share the same index. It's a lot like a table in a spreadsheet.
Below we create a DataFrame.
# Data: Air Quality Index (AQI) levels and temperature in different cities
data = {
"AQI": [55, 75, 60, 80, np.nan],
"Temperature": [68, 75, 64, 80, 85]
}
# Creating the DataFrame
df = pd.DataFrame(data, index = ["New York", "Los Angeles", "Chicago", "Houston", "Phoenix"])
df
| AQI | Temperature | |
|---|---|---|
| New York | 55.0 | 68 |
| Los Angeles | 75.0 | 75 |
| Chicago | 60.0 | 64 |
| Houston | 80.0 | 80 |
| Phoenix | NaN | 85 |
# You can set the style of the table
df.style.highlight_max()
| AQI | Temperature | |
|---|---|---|
| New York | 55.000000 | 68 |
| Los Angeles | 75.000000 | 75 |
| Chicago | 60.000000 | 64 |
| Houston | 80.000000 | 80 |
| Phoenix | nan | 85 |
You may notice that Pandas handles missing data very elegantly, keeping track of it through all calculations.
1.5 Index with Pandas DataFrame:¶
We can get a single column as a Series using python's getitem syntax on the DataFrame object.
df['AQI']
New York 55.0 Los Angeles 75.0 Chicago 60.0 Houston 80.0 Phoenix NaN Name: AQI, dtype: float64
...or using attribute syntax.
df.AQI
New York 55.0 Los Angeles 75.0 Chicago 60.0 Houston 80.0 Phoenix NaN Name: AQI, dtype: float64
If you want to get a specific row, use .loc
# label-based index: select the row whose index name is 'New York'
df.loc['New York']
AQI 55.0 Temperature 68.0 Name: New York, dtype: float64
# position-based index: Select the row at position 2 (the third row, since counting starts at 0).
df.iloc[2]
AQI 60.0 Temperature 64.0 Name: Chicago, dtype: float64
But we can also specify the column and row we want to access:
df.loc['New York','AQI']
np.float64(55.0)
Exercise 3: Print out the temperature of Chicago and Los Angeles. (3 points)¶
1.6 Basic statistics with Pandas:¶
# By default, this function works column-wise (axis=0), so you get one minimum value per column.
df.min()
AQI 55.0 Temperature 64.0 dtype: float64
# Set axis = 1, and you get the minimum value per row.
df.min(axis = 1)
New York 55.0 Los Angeles 75.0 Chicago 60.0 Houston 80.0 Phoenix 85.0 dtype: float64
# Return which row (city) has the max in each column (tempature or AQI).
df.idxmax()
AQI Houston Temperature Phoenix dtype: object
# Return which variable (tempature or AQI) has the max in each city.
df.idxmax(axis = 1)
New York Temperature Los Angeles AQI Chicago Temperature Houston AQI Phoenix Temperature dtype: object
# Return the mean of tempature and AQI
df.mean()
AQI 67.5 Temperature 74.4 dtype: float64
# Return the mean of specified variable.
df['AQI'].mean()
np.float64(67.5)
# Count the number of non-missing values (non-NaN) in each column
df.count()
AQI 4 Temperature 5 dtype: int64
# Standard deviation
df.std()
AQI 11.902381 Temperature 8.561542 dtype: float64
# describe function can print out the descriptive statistics easily!
df.describe()
| AQI | Temperature | |
|---|---|---|
| count | 4.000000 | 5.000000 |
| mean | 67.500000 | 74.400000 |
| std | 11.902381 | 8.561542 |
| min | 55.000000 | 64.000000 |
| 25% | 58.750000 | 68.000000 |
| 50% | 67.500000 | 75.000000 |
| 75% | 76.250000 | 80.000000 |
| max | 80.000000 | 85.000000 |
Exercise 4: Print the city name with the lowest temperature (3 points)¶
Note: Output only the city name. Points will be deducted if you include any extra or irrelevant information.
1.7 Modifying Pandas DataFrame¶
Add new row:
df.loc['New Brunswick'] = [64, 75]
df
| AQI | Temperature | |
|---|---|---|
| New York | 55.0 | 68 |
| Los Angeles | 75.0 | 75 |
| Chicago | 60.0 | 64 |
| Houston | 80.0 | 80 |
| Phoenix | NaN | 85 |
| New Brunswick | 64.0 | 75 |
Add new column:
df['Cloudy'] = [True, False, True, False, False, True]
df
| AQI | Temperature | Cloudy | |
|---|---|---|---|
| New York | 55.0 | 68 | True |
| Los Angeles | 75.0 | 75 | False |
| Chicago | 60.0 | 64 | True |
| Houston | 80.0 | 80 | False |
| Phoenix | NaN | 85 | False |
| New Brunswick | 64.0 | 75 | True |
data = {
"Latitude": [40.7128, 34.0522, 41.8781, 29.7604, 33.4484],
"Longitude": [-74.0060, -118.2437, -87.6298, -95.3698, -112.0740]
}
df_loc = pd.DataFrame(data, index = ["New York", "Los Angeles", "Chicago", "Houston", "Phoenix"])
df_loc
| Latitude | Longitude | |
|---|---|---|
| New York | 40.7128 | -74.0060 |
| Los Angeles | 34.0522 | -118.2437 |
| Chicago | 41.8781 | -87.6298 |
| Houston | 29.7604 | -95.3698 |
| Phoenix | 33.4484 | -112.0740 |
Combine df and df_loc side by side (column-wise) by matching their index labels.
df_join = df.join(df_loc)
df_join
| AQI | Temperature | Cloudy | Latitude | Longitude | |
|---|---|---|---|---|---|
| New York | 55.0 | 68 | True | 40.7128 | -74.0060 |
| Los Angeles | 75.0 | 75 | False | 34.0522 | -118.2437 |
| Chicago | 60.0 | 64 | True | 41.8781 | -87.6298 |
| Houston | 80.0 | 80 | False | 29.7604 | -95.3698 |
| Phoenix | NaN | 85 | False | 33.4484 | -112.0740 |
| New Brunswick | 64.0 | 75 | True | NaN | NaN |
# keep all rows from df_loc.
df_join = df.join(df_loc, how = 'right')
df_join
| AQI | Temperature | Cloudy | Latitude | Longitude | |
|---|---|---|---|---|---|
| New York | 55.0 | 68 | True | 40.7128 | -74.0060 |
| Los Angeles | 75.0 | 75 | False | 34.0522 | -118.2437 |
| Chicago | 60.0 | 64 | True | 41.8781 | -87.6298 |
| Houston | 80.0 | 80 | False | 29.7604 | -95.3698 |
| Phoenix | NaN | 85 | False | 33.4484 | -112.0740 |
# keep only rows with indexes in both.
df_join = df.join(df_loc, how = 'inner')
df_join
| AQI | Temperature | Cloudy | Latitude | Longitude | |
|---|---|---|---|---|---|
| New York | 55.0 | 68 | True | 40.7128 | -74.0060 |
| Los Angeles | 75.0 | 75 | False | 34.0522 | -118.2437 |
| Chicago | 60.0 | 64 | True | 41.8781 | -87.6298 |
| Houston | 80.0 | 80 | False | 29.7604 | -95.3698 |
| Phoenix | NaN | 85 | False | 33.4484 | -112.0740 |
# keep all rows from both, filling with NaN if needed
df_join = df.join(df_loc, how = 'outer')
df_join
| AQI | Temperature | Cloudy | Latitude | Longitude | |
|---|---|---|---|---|---|
| Chicago | 60.0 | 64 | True | 41.8781 | -87.6298 |
| Houston | 80.0 | 80 | False | 29.7604 | -95.3698 |
| Los Angeles | 75.0 | 75 | False | 34.0522 | -118.2437 |
| New Brunswick | 64.0 | 75 | True | NaN | NaN |
| New York | 55.0 | 68 | True | 40.7128 | -74.0060 |
| Phoenix | NaN | 85 | False | 33.4484 | -112.0740 |
Using concat to append new rows:
# glues together DataFrames along a particular axis. By default, appends df below another.
df_append = pd.concat([df,df_loc])
df_append
| AQI | Temperature | Cloudy | Latitude | Longitude | |
|---|---|---|---|---|---|
| New York | 55.0 | 68.0 | True | NaN | NaN |
| Los Angeles | 75.0 | 75.0 | False | NaN | NaN |
| Chicago | 60.0 | 64.0 | True | NaN | NaN |
| Houston | 80.0 | 80.0 | False | NaN | NaN |
| Phoenix | NaN | 85.0 | False | NaN | NaN |
| New Brunswick | 64.0 | 75.0 | True | NaN | NaN |
| New York | NaN | NaN | NaN | 40.7128 | -74.0060 |
| Los Angeles | NaN | NaN | NaN | 34.0522 | -118.2437 |
| Chicago | NaN | NaN | NaN | 41.8781 | -87.6298 |
| Houston | NaN | NaN | NaN | 29.7604 | -95.3698 |
| Phoenix | NaN | NaN | NaN | 33.4484 | -112.0740 |
Exercise 5: Combine New Jersey City Temperature and County Data (8 points)¶
Create two DataFrames (4 points):
df_temp: use city names as the index ("Newark","New Brunswick","Trenton").- Add one column
"Temperature". - Check today’s weather report (online or with a weather app) and fill in the current temperature values for each city.
- Add one column
df_county: use the same city names as the index and one column"County"with values["Essex", "Middlesex", "Mercer"].
Using
.join(): Merge the two DataFrames so the result shows each city’s temperature and county side by side (4 points)
As you can see, pd.read_csv() has quite a few parameters. Don't be overwhelmed – most of these are optional arguments that allow you to specify exactly how your data file is structured and which part(s) you want to import. In particular, the sep parameter allows the user to specify the type of delimiter used in the file. The default is a comma, but you can actually pass other common delimiters (such as sep='\t', which is a tab) to import other delimited files. The only required argument is a string specifying the filepath of your file.
pd.read_csv?
Signature: pd.read_csv( filepath_or_buffer: 'FilePath | ReadCsvBuffer[bytes] | ReadCsvBuffer[str]', *, sep: 'str | None | lib.NoDefault' = <no_default>, delimiter: 'str | None | lib.NoDefault' = None, header: "int | Sequence[int] | None | Literal['infer']" = 'infer', names: 'Sequence[Hashable] | None | lib.NoDefault' = <no_default>, index_col: 'IndexLabel | Literal[False] | None' = None, usecols: 'UsecolsArgType' = None, dtype: 'DtypeArg | None' = None, engine: 'CSVEngine | None' = None, converters: 'Mapping[Hashable, Callable] | None' = None, true_values: 'list | None' = None, false_values: 'list | None' = None, skipinitialspace: 'bool' = False, skiprows: 'list[int] | int | Callable[[Hashable], bool] | None' = None, skipfooter: 'int' = 0, nrows: 'int | None' = None, na_values: 'Hashable | Iterable[Hashable] | Mapping[Hashable, Iterable[Hashable]] | None' = None, keep_default_na: 'bool' = True, na_filter: 'bool' = True, verbose: 'bool | lib.NoDefault' = <no_default>, skip_blank_lines: 'bool' = True, parse_dates: 'bool | Sequence[Hashable] | None' = None, infer_datetime_format: 'bool | lib.NoDefault' = <no_default>, keep_date_col: 'bool | lib.NoDefault' = <no_default>, date_parser: 'Callable | lib.NoDefault' = <no_default>, date_format: 'str | dict[Hashable, str] | None' = None, dayfirst: 'bool' = False, cache_dates: 'bool' = True, iterator: 'bool' = False, chunksize: 'int | None' = None, compression: 'CompressionOptions' = 'infer', thousands: 'str | None' = None, decimal: 'str' = '.', lineterminator: 'str | None' = None, quotechar: 'str' = '"', quoting: 'int' = 0, doublequote: 'bool' = True, escapechar: 'str | None' = None, comment: 'str | None' = None, encoding: 'str | None' = None, encoding_errors: 'str | None' = 'strict', dialect: 'str | csv.Dialect | None' = None, on_bad_lines: 'str' = 'error', delim_whitespace: 'bool | lib.NoDefault' = <no_default>, low_memory: 'bool' = True, memory_map: 'bool' = False, float_precision: "Literal['high', 'legacy'] | None" = None, storage_options: 'StorageOptions | None' = None, dtype_backend: 'DtypeBackend | lib.NoDefault' = <no_default>, ) -> 'DataFrame | TextFileReader' Docstring: Read a comma-separated values (csv) file into DataFrame. Also supports optionally iterating or breaking of the file into chunks. Additional help can be found in the online docs for `IO Tools <https://pandas.pydata.org/pandas-docs/stable/user_guide/io.html>`_. Parameters ---------- filepath_or_buffer : str, path object or file-like object Any valid string path is acceptable. The string could be a URL. Valid URL schemes include http, ftp, s3, gs, and file. For file URLs, a host is expected. A local file could be: file://localhost/path/to/table.csv. If you want to pass in a path object, pandas accepts any ``os.PathLike``. By file-like object, we refer to objects with a ``read()`` method, such as a file handle (e.g. via builtin ``open`` function) or ``StringIO``. sep : str, default ',' Character or regex pattern to treat as the delimiter. If ``sep=None``, the C engine cannot automatically detect the separator, but the Python parsing engine can, meaning the latter will be used and automatically detect the separator from only the first valid row of the file by Python's builtin sniffer tool, ``csv.Sniffer``. In addition, separators longer than 1 character and different from ``'\s+'`` will be interpreted as regular expressions and will also force the use of the Python parsing engine. Note that regex delimiters are prone to ignoring quoted data. Regex example: ``'\r\t'``. delimiter : str, optional Alias for ``sep``. header : int, Sequence of int, 'infer' or None, default 'infer' Row number(s) containing column labels and marking the start of the data (zero-indexed). Default behavior is to infer the column names: if no ``names`` are passed the behavior is identical to ``header=0`` and column names are inferred from the first line of the file, if column names are passed explicitly to ``names`` then the behavior is identical to ``header=None``. Explicitly pass ``header=0`` to be able to replace existing names. The header can be a list of integers that specify row locations for a :class:`~pandas.MultiIndex` on the columns e.g. ``[0, 1, 3]``. Intervening rows that are not specified will be skipped (e.g. 2 in this example is skipped). Note that this parameter ignores commented lines and empty lines if ``skip_blank_lines=True``, so ``header=0`` denotes the first line of data rather than the first line of the file. names : Sequence of Hashable, optional Sequence of column labels to apply. If the file contains a header row, then you should explicitly pass ``header=0`` to override the column names. Duplicates in this list are not allowed. index_col : Hashable, Sequence of Hashable or False, optional Column(s) to use as row label(s), denoted either by column labels or column indices. If a sequence of labels or indices is given, :class:`~pandas.MultiIndex` will be formed for the row labels. Note: ``index_col=False`` can be used to force pandas to *not* use the first column as the index, e.g., when you have a malformed file with delimiters at the end of each line. usecols : Sequence of Hashable or Callable, optional Subset of columns to select, denoted either by column labels or column indices. If list-like, all elements must either be positional (i.e. integer indices into the document columns) or strings that correspond to column names provided either by the user in ``names`` or inferred from the document header row(s). If ``names`` are given, the document header row(s) are not taken into account. For example, a valid list-like ``usecols`` parameter would be ``[0, 1, 2]`` or ``['foo', 'bar', 'baz']``. Element order is ignored, so ``usecols=[0, 1]`` is the same as ``[1, 0]``. To instantiate a :class:`~pandas.DataFrame` from ``data`` with element order preserved use ``pd.read_csv(data, usecols=['foo', 'bar'])[['foo', 'bar']]`` for columns in ``['foo', 'bar']`` order or ``pd.read_csv(data, usecols=['foo', 'bar'])[['bar', 'foo']]`` for ``['bar', 'foo']`` order. If callable, the callable function will be evaluated against the column names, returning names where the callable function evaluates to ``True``. An example of a valid callable argument would be ``lambda x: x.upper() in ['AAA', 'BBB', 'DDD']``. Using this parameter results in much faster parsing time and lower memory usage. dtype : dtype or dict of {Hashable : dtype}, optional Data type(s) to apply to either the whole dataset or individual columns. E.g., ``{'a': np.float64, 'b': np.int32, 'c': 'Int64'}`` Use ``str`` or ``object`` together with suitable ``na_values`` settings to preserve and not interpret ``dtype``. If ``converters`` are specified, they will be applied INSTEAD of ``dtype`` conversion. .. versionadded:: 1.5.0 Support for ``defaultdict`` was added. Specify a ``defaultdict`` as input where the default determines the ``dtype`` of the columns which are not explicitly listed. engine : {'c', 'python', 'pyarrow'}, optional Parser engine to use. The C and pyarrow engines are faster, while the python engine is currently more feature-complete. Multithreading is currently only supported by the pyarrow engine. .. versionadded:: 1.4.0 The 'pyarrow' engine was added as an *experimental* engine, and some features are unsupported, or may not work correctly, with this engine. converters : dict of {Hashable : Callable}, optional Functions for converting values in specified columns. Keys can either be column labels or column indices. true_values : list, optional Values to consider as ``True`` in addition to case-insensitive variants of 'True'. false_values : list, optional Values to consider as ``False`` in addition to case-insensitive variants of 'False'. skipinitialspace : bool, default False Skip spaces after delimiter. skiprows : int, list of int or Callable, optional Line numbers to skip (0-indexed) or number of lines to skip (``int``) at the start of the file. If callable, the callable function will be evaluated against the row indices, returning ``True`` if the row should be skipped and ``False`` otherwise. An example of a valid callable argument would be ``lambda x: x in [0, 2]``. skipfooter : int, default 0 Number of lines at bottom of file to skip (Unsupported with ``engine='c'``). nrows : int, optional Number of rows of file to read. Useful for reading pieces of large files. na_values : Hashable, Iterable of Hashable or dict of {Hashable : Iterable}, optional Additional strings to recognize as ``NA``/``NaN``. If ``dict`` passed, specific per-column ``NA`` values. By default the following values are interpreted as ``NaN``: " ", "#N/A", "#N/A N/A", "#NA", "-1.#IND", "-1.#QNAN", "-NaN", "-nan", "1.#IND", "1.#QNAN", "<NA>", "N/A", "NA", "NULL", "NaN", "None", "n/a", "nan", "null ". keep_default_na : bool, default True Whether or not to include the default ``NaN`` values when parsing the data. Depending on whether ``na_values`` is passed in, the behavior is as follows: * If ``keep_default_na`` is ``True``, and ``na_values`` are specified, ``na_values`` is appended to the default ``NaN`` values used for parsing. * If ``keep_default_na`` is ``True``, and ``na_values`` are not specified, only the default ``NaN`` values are used for parsing. * If ``keep_default_na`` is ``False``, and ``na_values`` are specified, only the ``NaN`` values specified ``na_values`` are used for parsing. * If ``keep_default_na`` is ``False``, and ``na_values`` are not specified, no strings will be parsed as ``NaN``. Note that if ``na_filter`` is passed in as ``False``, the ``keep_default_na`` and ``na_values`` parameters will be ignored. na_filter : bool, default True Detect missing value markers (empty strings and the value of ``na_values``). In data without any ``NA`` values, passing ``na_filter=False`` can improve the performance of reading a large file. verbose : bool, default False Indicate number of ``NA`` values placed in non-numeric columns. .. deprecated:: 2.2.0 skip_blank_lines : bool, default True If ``True``, skip over blank lines rather than interpreting as ``NaN`` values. parse_dates : bool, list of Hashable, list of lists or dict of {Hashable : list}, default False The behavior is as follows: * ``bool``. If ``True`` -> try parsing the index. Note: Automatically set to ``True`` if ``date_format`` or ``date_parser`` arguments have been passed. * ``list`` of ``int`` or names. e.g. If ``[1, 2, 3]`` -> try parsing columns 1, 2, 3 each as a separate date column. * ``list`` of ``list``. e.g. If ``[[1, 3]]`` -> combine columns 1 and 3 and parse as a single date column. Values are joined with a space before parsing. * ``dict``, e.g. ``{'foo' : [1, 3]}`` -> parse columns 1, 3 as date and call result 'foo'. Values are joined with a space before parsing. If a column or index cannot be represented as an array of ``datetime``, say because of an unparsable value or a mixture of timezones, the column or index will be returned unaltered as an ``object`` data type. For non-standard ``datetime`` parsing, use :func:`~pandas.to_datetime` after :func:`~pandas.read_csv`. Note: A fast-path exists for iso8601-formatted dates. infer_datetime_format : bool, default False If ``True`` and ``parse_dates`` is enabled, pandas will attempt to infer the format of the ``datetime`` strings in the columns, and if it can be inferred, switch to a faster method of parsing them. In some cases this can increase the parsing speed by 5-10x. .. deprecated:: 2.0.0 A strict version of this argument is now the default, passing it has no effect. keep_date_col : bool, default False If ``True`` and ``parse_dates`` specifies combining multiple columns then keep the original columns. date_parser : Callable, optional Function to use for converting a sequence of string columns to an array of ``datetime`` instances. The default uses ``dateutil.parser.parser`` to do the conversion. pandas will try to call ``date_parser`` in three different ways, advancing to the next if an exception occurs: 1) Pass one or more arrays (as defined by ``parse_dates``) as arguments; 2) concatenate (row-wise) the string values from the columns defined by ``parse_dates`` into a single array and pass that; and 3) call ``date_parser`` once for each row using one or more strings (corresponding to the columns defined by ``parse_dates``) as arguments. .. deprecated:: 2.0.0 Use ``date_format`` instead, or read in as ``object`` and then apply :func:`~pandas.to_datetime` as-needed. date_format : str or dict of column -> format, optional Format to use for parsing dates when used in conjunction with ``parse_dates``. The strftime to parse time, e.g. :const:`"%d/%m/%Y"`. See `strftime documentation <https://docs.python.org/3/library/datetime.html #strftime-and-strptime-behavior>`_ for more information on choices, though note that :const:`"%f"` will parse all the way up to nanoseconds. You can also pass: - "ISO8601", to parse any `ISO8601 <https://en.wikipedia.org/wiki/ISO_8601>`_ time string (not necessarily in exactly the same format); - "mixed", to infer the format for each element individually. This is risky, and you should probably use it along with `dayfirst`. .. versionadded:: 2.0.0 dayfirst : bool, default False DD/MM format dates, international and European format. cache_dates : bool, default True If ``True``, use a cache of unique, converted dates to apply the ``datetime`` conversion. May produce significant speed-up when parsing duplicate date strings, especially ones with timezone offsets. iterator : bool, default False Return ``TextFileReader`` object for iteration or getting chunks with ``get_chunk()``. chunksize : int, optional Number of lines to read from the file per chunk. Passing a value will cause the function to return a ``TextFileReader`` object for iteration. See the `IO Tools docs <https://pandas.pydata.org/pandas-docs/stable/io.html#io-chunking>`_ for more information on ``iterator`` and ``chunksize``. compression : str or dict, default 'infer' For on-the-fly decompression of on-disk data. If 'infer' and 'filepath_or_buffer' is path-like, then detect compression from the following extensions: '.gz', '.bz2', '.zip', '.xz', '.zst', '.tar', '.tar.gz', '.tar.xz' or '.tar.bz2' (otherwise no compression). If using 'zip' or 'tar', the ZIP file must contain only one data file to be read in. Set to ``None`` for no decompression. Can also be a dict with key ``'method'`` set to one of {``'zip'``, ``'gzip'``, ``'bz2'``, ``'zstd'``, ``'xz'``, ``'tar'``} and other key-value pairs are forwarded to ``zipfile.ZipFile``, ``gzip.GzipFile``, ``bz2.BZ2File``, ``zstandard.ZstdDecompressor``, ``lzma.LZMAFile`` or ``tarfile.TarFile``, respectively. As an example, the following could be passed for Zstandard decompression using a custom compression dictionary: ``compression={'method': 'zstd', 'dict_data': my_compression_dict}``. .. versionadded:: 1.5.0 Added support for `.tar` files. .. versionchanged:: 1.4.0 Zstandard support. thousands : str (length 1), optional Character acting as the thousands separator in numerical values. decimal : str (length 1), default '.' Character to recognize as decimal point (e.g., use ',' for European data). lineterminator : str (length 1), optional Character used to denote a line break. Only valid with C parser. quotechar : str (length 1), optional Character used to denote the start and end of a quoted item. Quoted items can include the ``delimiter`` and it will be ignored. quoting : {0 or csv.QUOTE_MINIMAL, 1 or csv.QUOTE_ALL, 2 or csv.QUOTE_NONNUMERIC, 3 or csv.QUOTE_NONE}, default csv.QUOTE_MINIMAL Control field quoting behavior per ``csv.QUOTE_*`` constants. Default is ``csv.QUOTE_MINIMAL`` (i.e., 0) which implies that only fields containing special characters are quoted (e.g., characters defined in ``quotechar``, ``delimiter``, or ``lineterminator``. doublequote : bool, default True When ``quotechar`` is specified and ``quoting`` is not ``QUOTE_NONE``, indicate whether or not to interpret two consecutive ``quotechar`` elements INSIDE a field as a single ``quotechar`` element. escapechar : str (length 1), optional Character used to escape other characters. comment : str (length 1), optional Character indicating that the remainder of line should not be parsed. If found at the beginning of a line, the line will be ignored altogether. This parameter must be a single character. Like empty lines (as long as ``skip_blank_lines=True``), fully commented lines are ignored by the parameter ``header`` but not by ``skiprows``. For example, if ``comment='#'``, parsing ``#empty\na,b,c\n1,2,3`` with ``header=0`` will result in ``'a,b,c'`` being treated as the header. encoding : str, optional, default 'utf-8' Encoding to use for UTF when reading/writing (ex. ``'utf-8'``). `List of Python standard encodings <https://docs.python.org/3/library/codecs.html#standard-encodings>`_ . encoding_errors : str, optional, default 'strict' How encoding errors are treated. `List of possible values <https://docs.python.org/3/library/codecs.html#error-handlers>`_ . .. versionadded:: 1.3.0 dialect : str or csv.Dialect, optional If provided, this parameter will override values (default or not) for the following parameters: ``delimiter``, ``doublequote``, ``escapechar``, ``skipinitialspace``, ``quotechar``, and ``quoting``. If it is necessary to override values, a ``ParserWarning`` will be issued. See ``csv.Dialect`` documentation for more details. on_bad_lines : {'error', 'warn', 'skip'} or Callable, default 'error' Specifies what to do upon encountering a bad line (a line with too many fields). Allowed values are : - ``'error'``, raise an Exception when a bad line is encountered. - ``'warn'``, raise a warning when a bad line is encountered and skip that line. - ``'skip'``, skip bad lines without raising or warning when they are encountered. .. versionadded:: 1.3.0 .. versionadded:: 1.4.0 - Callable, function with signature ``(bad_line: list[str]) -> list[str] | None`` that will process a single bad line. ``bad_line`` is a list of strings split by the ``sep``. If the function returns ``None``, the bad line will be ignored. If the function returns a new ``list`` of strings with more elements than expected, a ``ParserWarning`` will be emitted while dropping extra elements. Only supported when ``engine='python'`` .. versionchanged:: 2.2.0 - Callable, function with signature as described in `pyarrow documentation <https://arrow.apache.org/docs/python/generated/pyarrow.csv.ParseOptions.html #pyarrow.csv.ParseOptions.invalid_row_handler>`_ when ``engine='pyarrow'`` delim_whitespace : bool, default False Specifies whether or not whitespace (e.g. ``' '`` or ``'\t'``) will be used as the ``sep`` delimiter. Equivalent to setting ``sep='\s+'``. If this option is set to ``True``, nothing should be passed in for the ``delimiter`` parameter. .. deprecated:: 2.2.0 Use ``sep="\s+"`` instead. low_memory : bool, default True Internally process the file in chunks, resulting in lower memory use while parsing, but possibly mixed type inference. To ensure no mixed types either set ``False``, or specify the type with the ``dtype`` parameter. Note that the entire file is read into a single :class:`~pandas.DataFrame` regardless, use the ``chunksize`` or ``iterator`` parameter to return the data in chunks. (Only valid with C parser). memory_map : bool, default False If a filepath is provided for ``filepath_or_buffer``, map the file object directly onto memory and access the data directly from there. Using this option can improve performance because there is no longer any I/O overhead. float_precision : {'high', 'legacy', 'round_trip'}, optional Specifies which converter the C engine should use for floating-point values. The options are ``None`` or ``'high'`` for the ordinary converter, ``'legacy'`` for the original lower precision pandas converter, and ``'round_trip'`` for the round-trip converter. storage_options : dict, optional Extra options that make sense for a particular storage connection, e.g. host, port, username, password, etc. For HTTP(S) URLs the key-value pairs are forwarded to ``urllib.request.Request`` as header options. For other URLs (e.g. starting with "s3://", and "gcs://") the key-value pairs are forwarded to ``fsspec.open``. Please see ``fsspec`` and ``urllib`` for more details, and for more examples on storage options refer `here <https://pandas.pydata.org/docs/user_guide/io.html? highlight=storage_options#reading-writing-remote-files>`_. dtype_backend : {'numpy_nullable', 'pyarrow'}, default 'numpy_nullable' Back-end data type applied to the resultant :class:`DataFrame` (still experimental). Behaviour is as follows: * ``"numpy_nullable"``: returns nullable-dtype-backed :class:`DataFrame` (default). * ``"pyarrow"``: returns pyarrow-backed nullable :class:`ArrowDtype` DataFrame. .. versionadded:: 2.0 Returns ------- DataFrame or TextFileReader A comma-separated values (csv) file is returned as two-dimensional data structure with labeled axes. See Also -------- DataFrame.to_csv : Write DataFrame to a comma-separated values (csv) file. read_table : Read general delimited file into DataFrame. read_fwf : Read a table of fixed-width formatted lines into DataFrame. Examples -------- >>> pd.read_csv('data.csv') # doctest: +SKIP File: ~/miniforge3/envs/esa_env/lib/python3.9/site-packages/pandas/io/parsers/readers.py Type: function
Download wildfire data Spatial_Database_Big_Wildfires_US_all.csv from Canvas, and upload the data to your current working directory.
df = pd.read_csv('Spatial_Database_Big_Wildfires_US_all.csv')
df.head()
| FOD_ID | FPA_ID | SOURCE_SYSTEM_TYPE | SOURCE_SYSTEM | NWCG_REPORTING_UNIT_ID | NWCG_REPORTING_UNIT_NAME | FIRE_CODE | FIRE_NAME | MTBS_FIRE_NAME | COMPLEX_NAME | ... | CONT_DOY | FIRE_SIZE | FIRE_SIZE_CLASS | LATITUDE | LONGITUDE | OWNER_DESCR | STATE | COUNTY | FIPS_CODE | FIPS_NAME | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17 | FS-1418878 | FED | FS-FIRESTAT | USCAENF | Eldorado National Forest | NaN | POWER | POWER | NaN | ... | 295.0 | 16823.0 | G | 38.523333 | -120.211667 | USFS | CA | 5 | 6005.0 | Amador County |
| 1 | 18 | FS-1418881 | FED | FS-FIRESTAT | USCAENF | Eldorado National Forest | BHA3 | FREDS | FREDS | NaN | ... | 291.0 | 7700.0 | G | 38.780000 | -120.260000 | USFS | CA | 17 | 6017.0 | El Dorado County |
| 2 | 40 | FS-1418920 | FED | FS-FIRESTAT | USNCNCF | National Forests in North Carolina | BKC8 | AUSTIN CREEK | NaN | NaN | ... | 44.0 | 125.0 | D | 36.001667 | -81.590000 | MISSING/NOT SPECIFIED | NC | 27 | 37027.0 | Caldwell County |
| 3 | 119 | FS-1419153 | FED | FS-FIRESTAT | USNENBF | Nebraska National Forest | BEW8 | THOMPSON BUTTE | NaN | NaN | ... | 198.0 | 119.0 | D | 43.899167 | -102.954722 | USFS | SD | 103 | 46103.0 | Pennington County |
| 4 | 120 | FS-1419156 | FED | FS-FIRESTAT | USNENBF | Nebraska National Forest | BEW8 | CHARLES DRAW | NaN | NaN | ... | 197.0 | 119.0 | D | 43.892778 | -102.948056 | USFS | SD | 103 | 46103.0 | Pennington County |
5 rows × 28 columns
2.2 Groupby methods¶
groupby is an amazingly powerful function in pandas, but it is also complicated to use and understand.
The point of this section is to make you feel confident in using groupby.
df.columns
Index(['FOD_ID', 'FPA_ID', 'SOURCE_SYSTEM_TYPE', 'SOURCE_SYSTEM',
'NWCG_REPORTING_UNIT_ID', 'NWCG_REPORTING_UNIT_NAME', 'FIRE_CODE',
'FIRE_NAME', 'MTBS_FIRE_NAME', 'COMPLEX_NAME', 'FIRE_YEAR',
'DISCOVERY_DATE', 'DISCOVERY_DOY', 'DISCOVERY_TIME',
'NWCG_CAUSE_CLASSIFICATION', 'NWCG_GENERAL_CAUSE',
'NWCG_CAUSE_AGE_CATEGORY', 'CONT_DATE', 'CONT_DOY', 'FIRE_SIZE',
'FIRE_SIZE_CLASS', 'LATITUDE', 'LONGITUDE', 'OWNER_DESCR', 'STATE',
'COUNTY', 'FIPS_CODE', 'FIPS_NAME'],
dtype='object')
df = df.set_index('FOD_ID') #Set index
df[['DISCOVERY_DATE','DISCOVERY_DOY','DISCOVERY_TIME' ]]
| DISCOVERY_DATE | DISCOVERY_DOY | DISCOVERY_TIME | |
|---|---|---|---|
| FOD_ID | |||
| 17 | 10/6/2004 | 280 | 1415.0 |
| 18 | 10/13/2004 | 287 | 1618.0 |
| 40 | 2/12/2005 | 43 | 1520.0 |
| 119 | 7/16/2005 | 197 | 1715.0 |
| 120 | 7/16/2005 | 197 | 1730.0 |
| ... | ... | ... | ... |
| 400732975 | 8/9/2019 | 221 | 2134.0 |
| 400732976 | 3/1/2020 | 61 | 1330.0 |
| 400732977 | 5/13/2020 | 134 | 1300.0 |
| 400732982 | 8/17/2020 | 230 | 755.0 |
| 400732984 | 11/20/2020 | 325 | 1110.0 |
60713 rows × 3 columns
An Example:¶
Question: Find out the top 10 states with largest number of wildfires.
This is an example of a "one-liner" that you can accomplish with groupby.
df.groupby('STATE').FPA_ID.count().nlargest(10).plot(kind='bar', figsize=(12,6))
<Axes: xlabel='STATE'>
What Happened?¶
Let's break apart this operation a bit. The workflow with groupby can be divided into three general steps:
Split: Partition the data into different groups based on some criterion.
Apply: Do some caclulation within each group. Different types of "apply" steps might be
- Aggregation: Get the mean or max within the group.
- Transformation: Normalize all the values within a group.
- Filtration: Eliminate some groups based on a criterion.
Combine: Put the results back together into a single object.
The groupby method¶
Both Series and DataFrame objects have a groupby method. It accepts a variety of arguments, but the simplest way to think about it is that you pass another series, whose unique values are used to split the original object into different groups.
# Group by the STATE.
df.groupby(df.STATE)
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x15bb54370>
There is a shortcut for doing this with dataframes: you just pass the column name:
df.groupby('STATE')
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x15bb54340>
The GroubBy object¶
When we call, groupby we get back a GroupBy object:
gb = df.groupby('STATE')
gb
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x15bb549a0>
The length tells us how many groups were found:
len(gb)
50
All of the groups are available as a dictionary via the .groups attribute:
groups = gb.groups
len(groups)
50
# Access group 'NJ'
groups['NJ']
Index([ 247140, 384407, 578890, 579164, 579296, 580135,
580956, 581165, 582330, 582680,
...
400271442, 400389794, 400482043, 400528139, 400531379, 400587134,
400594776, 400602511, 400613352, 400632920],
dtype='int64', name='FOD_ID', length=111)
2.3 Iterating and selecting groups¶
You can loop through the groups if you want.
for key, group in gb:
display(group.head())
print(f'The key is "{key}"')
break
| FPA_ID | SOURCE_SYSTEM_TYPE | SOURCE_SYSTEM | NWCG_REPORTING_UNIT_ID | NWCG_REPORTING_UNIT_NAME | FIRE_CODE | FIRE_NAME | MTBS_FIRE_NAME | COMPLEX_NAME | FIRE_YEAR | ... | CONT_DOY | FIRE_SIZE | FIRE_SIZE_CLASS | LATITUDE | LONGITUDE | OWNER_DESCR | STATE | COUNTY | FIPS_CODE | FIPS_NAME | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOD_ID | |||||||||||||||||||||
| 6689 | FS-1431539 | FED | FS-FIRESTAT | USAKTNF | Tongass National Forest | BRD1 | MUSKEG | NaN | NaN | 2005 | ... | 126.0 | 305.0 | E | 59.087222 | -135.441389 | STATE OR PRIVATE | AK | 220 | 2220.0 | Sitka City and Borough |
| 109456 | FS-334441 | FED | FS-FIRESTAT | USAKTNF | Tongass National Forest | NaN | MILL | NaN | NaN | 1998 | ... | 189.0 | 118.0 | D | 55.681667 | -132.615000 | STATE OR PRIVATE | AK | NaN | NaN | NaN |
| 147361 | FS-374211 | FED | FS-FIRESTAT | USAKCGF | Chugach National Forest | NaN | KENAI LAKE | KENAI LAKE | NaN | 2001 | ... | 188.0 | 3260.0 | F | 60.410278 | -149.473611 | USFS | AK | NaN | NaN | NaN |
| 174677 | W-374459 | FED | DOI-WFMI | USAKAKA | Alaska Regional Office | B391 | B391 | 532391 | NaN | 1995 | ... | 226.0 | 2850.0 | F | 66.832700 | -160.736100 | TRIBAL | AK | NaN | NaN | NaN |
| 213301 | W-36457 | FED | DOI-WFMI | USAKAKD | Alaska Fire Service | A029 | 203029 | NaN | NaN | 1992 | ... | 126.0 | 170.0 | D | 57.065900 | -154.085700 | BIA | AK | NaN | NaN | NaN |
5 rows × 27 columns
The key is "AK"
key→ the group label, i.e. the value from the"STATE"column (like"CA","TX","NJ").group→ the sub-DataFrame containing all the rows fromdfthat belong to that group.
And you can get a specific group by key.
gb.get_group('NJ')
| FPA_ID | SOURCE_SYSTEM_TYPE | SOURCE_SYSTEM | NWCG_REPORTING_UNIT_ID | NWCG_REPORTING_UNIT_NAME | FIRE_CODE | FIRE_NAME | MTBS_FIRE_NAME | COMPLEX_NAME | FIRE_YEAR | ... | CONT_DOY | FIRE_SIZE | FIRE_SIZE_CLASS | LATITUDE | LONGITUDE | OWNER_DESCR | STATE | COUNTY | FIPS_CODE | FIPS_NAME | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOD_ID | |||||||||||||||||||||
| 247140 | W-234848 | FED | DOI-WFMI | USPADWP | Delaware Water Gap National Recreation Area | NaN | WORTHINGTO | WORTHINGTO | NaN | 1999 | ... | 102.0 | 623.0 | E | 40.995895 | -75.120000 | NPS | NJ | NaN | NaN | NaN |
| 384407 | FWS-2007NJERRDFR2 | FED | FWS-FMIS | USNJERR | Edwin B. Forsythe National Wildlife Refuge | DFR2 | NJ NJFFS WF ASSIST WARREN GROVE | WARREN GROVE | NaN | 2007 | ... | 141.0 | 17050.0 | G | 39.707500 | -74.309722 | STATE | NJ | NaN | NaN | NaN |
| 578890 | SFO-2006NJDEPA032704 | NONFED | ST-NASF | USNJNJS | New Jersey Forest Fire Service | NaN | NaN | NaN | NaN | 2006 | ... | NaN | 104.0 | D | 40.304400 | -74.201100 | PRIVATE | NJ | Middlesex | 34023.0 | Middlesex County |
| 579164 | SFO-2006NJDEPB012703 | NONFED | ST-NASF | USNJNJS | New Jersey Forest Fire Service | NaN | RARITAN CENTER | NaN | NaN | 2006 | ... | NaN | 450.0 | E | 40.296100 | -74.214000 | PRIVATE | NJ | Middlesex | 34023.0 | Middlesex County |
| 579296 | SFO-2006NJDEPB032108 | NONFED | ST-NASF | USNJNJS | New Jersey Forest Fire Service | NaN | SUNRISE LAKE | NaN | NaN | 2006 | ... | NaN | 136.0 | D | 39.482800 | -74.510900 | STATE | NJ | Burlington | 34005.0 | Burlington County |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 400587134 | SFO-2020NJDEPA03-200223155509 | NONFED | ST-NASF | USNJNJS | New Jersey Forest Fire Service | NaN | NaN | NaN | NaN | 2020 | ... | NaN | 103.0 | D | 40.970480 | -75.117710 | MISSING/NOT SPECIFIED | NJ | Warren | 34041.0 | Warren County |
| 400594776 | SFO-2020NJDEPC03-200409234633 | NONFED | ST-NASF | USNJNJS | New Jersey Forest Fire Service | NaN | NaN | SPLIT DITCH | NaN | 2020 | ... | NaN | 1518.0 | F | 39.312640 | -75.090240 | MISSING/NOT SPECIFIED | NJ | Cumberland | 34011.0 | Cumberland County |
| 400602511 | SFO-2020NJDEPC06-200519235337 | NONFED | ST-NASF | USNJNJS | New Jersey Forest Fire Service | NaN | NaN | BIG TIMBER | NaN | 2020 | ... | NaN | 2107.0 | F | 39.651250 | -74.892050 | MISSING/NOT SPECIFIED | NJ | Camden | 34007.0 | Camden County |
| 400613352 | SFO-2020NJDEPB09-200709201959 | NONFED | ST-NASF | USNJNJS | New Jersey Forest Fire Service | NaN | NaN | NaN | NaN | 2020 | ... | NaN | 204.0 | D | 40.111570 | -74.412330 | MISSING/NOT SPECIFIED | NJ | Ocean | 34029.0 | Ocean County |
| 400632920 | ICS209_2019_10720324 | INTERAGCY | IA-ICS209 | USNJNJS | New Jersey Forest Fire Service | NaN | SPRING HILL FIRE | SPRING HILL FIRE | NaN | 2019 | ... | NaN | 11638.0 | G | 39.770000 | -74.450000 | MISSING/NOT SPECIFIED | NJ | Burlington | 34005.0 | Burlington County |
111 rows × 27 columns
2.4 Aggregation¶
Now that we know how to create a GroupBy object, let's learn how to do aggregation on it.
One way us to use the .aggregate method, which accepts another function as its argument. The result is automatically combined into a new dataframe with the group key as the index.
By default, the operation is applied to every column. That's usually not what we want. We can use both . or [] syntax to select a specific column to operate on. Then we get back a series.
# Find out the 10 states with biggest fire size.
gb.FIRE_SIZE.max().nlargest(10)
STATE OK 662700.0 AK 606945.0 CA 589368.0 OR 558198.3 AZ 538049.0 TX 479549.0 NV 416821.2 ID 367785.0 UT 357185.0 GA 309200.0 Name: FIRE_SIZE, dtype: float64
There are shortcuts for common aggregation functions:
gb.FIRE_SIZE.max().nlargest(10)
STATE OK 662700.0 AK 606945.0 CA 589368.0 OR 558198.3 AZ 538049.0 TX 479549.0 NV 416821.2 ID 367785.0 UT 357185.0 GA 309200.0 Name: FIRE_SIZE, dtype: float64
gb.FIRE_SIZE.mean().nlargest(10)
STATE AK 16440.375603 NV 6138.109191 OR 5595.310524 WA 5071.806130 ID 4657.836385 CA 4086.919845 MT 3571.759463 AZ 3084.376006 UT 2874.009960 CO 2809.130881 Name: FIRE_SIZE, dtype: float64
gb.FIRE_SIZE.sum().nlargest(10).plot(kind='bar')
<Axes: xlabel='STATE'>
# Find out unique values of fire cause classification.
df['NWCG_CAUSE_CLASSIFICATION'].unique()
array(['Human', 'Natural', 'Missing data/not specified/undetermined'],
dtype=object)
# Find out unique values of fire general cause.
df['NWCG_GENERAL_CAUSE'].unique()
array(['Equipment and vehicle use',
'Power generation/transmission/distribution',
'Debris and open burning', 'Natural',
'Missing data/not specified/undetermined',
'Recreation and ceremony', 'Smoking',
'Railroad operations and maintenance', 'Arson/incendiarism',
'Fireworks', 'Other causes', 'Misuse of fire by a minor',
'Firearms and explosives use'], dtype=object)
# Find out the 10 leading causes of fires.
df.groupby('NWCG_GENERAL_CAUSE').count()['FPA_ID'].nlargest(10).plot(kind = 'bar')
<Axes: xlabel='NWCG_GENERAL_CAUSE'>
Exercise 6: Plot the number of wildfires every year. (5 points)¶
Hint: Use groupby method. Try making this to be one line of code.
Exercise 7: Plot the number of wildfires in CA every year. (5 points)¶
2.5 Groupby multiple index¶
gb = df.groupby(['STATE','FIRE_YEAR'])
len(gb)
1231
list(gb.groups.keys())[:100:10]
[('AK', 1992),
('AK', 2002),
('AK', 2012),
('AL', 1993),
('AL', 2003),
('AL', 2013),
('AR', 1994),
('AR', 2004),
('AR', 2014),
('AZ', 1995)]
gb.FIRE_SIZE.sum()
STATE FIRE_YEAR
AK 1992 141007.000
1993 684669.800
1994 259901.600
1995 42526.000
1996 596706.400
...
WY 2016 254804.700
2017 118803.020
2018 220718.000
2019 41022.200
2020 285791.255
Name: FIRE_SIZE, Length: 1231, dtype: float64
### Select group with multiple index must use tuple!
gb.get_group(('CA', 2003))
| FPA_ID | SOURCE_SYSTEM_TYPE | SOURCE_SYSTEM | NWCG_REPORTING_UNIT_ID | NWCG_REPORTING_UNIT_NAME | FIRE_CODE | FIRE_NAME | MTBS_FIRE_NAME | COMPLEX_NAME | FIRE_YEAR | ... | CONT_DOY | FIRE_SIZE | FIRE_SIZE_CLASS | LATITUDE | LONGITUDE | OWNER_DESCR | STATE | COUNTY | FIPS_CODE | FIPS_NAME | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOD_ID | |||||||||||||||||||||
| 157551 | FS-385211 | FED | FS-FIRESTAT | USCAINF | Inyo National Forest | NaN | DEXTER | DEXTER WFU | NaN | 2003 | ... | 286.0 | 2515.0 | F | 37.831389 | -118.795000 | USFS | CA | NaN | NaN | NaN |
| 158728 | FS-386431 | FED | FS-FIRESTAT | USCAPNF | Plumas National Forest | 4300 | ROWLAND | NaN | NaN | 2003 | ... | 163.0 | 114.0 | D | 39.951389 | -120.068889 | USFS | CA | NaN | NaN | NaN |
| 159533 | FS-387254 | FED | FS-FIRESTAT | USCASTF | Stanislaus National Forest | 7648 | MUDD | MUD WFU | MUD COMPLEX | 2003 | ... | 300.0 | 4102.0 | F | 38.424722 | -119.961111 | USFS | CA | NaN | NaN | NaN |
| 159534 | FS-387255 | FED | FS-FIRESTAT | USCASTF | Stanislaus National Forest | 5555 | WHITT | WHITT | MUD COMPLEX | 2003 | ... | 300.0 | 1014.0 | F | 38.378056 | -119.999722 | USFS | CA | NaN | NaN | NaN |
| 160202 | FS-388122 | FED | FS-FIRESTAT | USCALPF | Los Padres National Forest | 2996 | DEL VENTURI | NaN | NaN | 2003 | ... | 225.0 | 861.0 | E | 36.071111 | -121.390000 | MISSING/NOT SPECIFIED | CA | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15000827 | ICS209_2003_CA-KRN-37559 | INTERAGCY | IA-ICS209 | USCAKRN | Kern County Fire Department | NaN | HILLSIDE | UNNAMED | NaN | 2003 | ... | 198.0 | 835.0 | E | 35.168056 | -118.468056 | MISSING/NOT SPECIFIED | CA | KERN | 6029.0 | Kern County |
| 15000828 | ICS209_2003_CA-KRN-38766 | INTERAGCY | IA-ICS209 | USCAKRN | Kern County Fire Department | NaN | MERCURY | UNNAMED | NaN | 2003 | ... | 203.0 | 340.0 | E | 35.200556 | -118.518611 | MISSING/NOT SPECIFIED | CA | KERN | 6029.0 | Kern County |
| 15000829 | ICS209_2003_CA-LAC-03004054 | INTERAGCY | IA-ICS209 | USCALAC | Los Angeles County Fire Department | NaN | AIRPORT | NaN | NaN | 2003 | ... | 8.0 | 245.0 | D | 33.407778 | -118.402500 | MISSING/NOT SPECIFIED | CA | LOS ANGELES | 6037.0 | Los Angeles County |
| 201940026 | ICS209_2003-CA-KRN-0333259 | INTERAGCY | IA-ICS209 | USCAKRN | Kern County Fire Department | NaN | TEJON | TEJON | NaN | 2003 | ... | 183.0 | 1155.0 | F | 34.871389 | -118.882778 | MISSING/NOT SPECIFIED | CA | Kern | 6029.0 | Kern County |
| 400280041 | ICS209_2003_CA-KRN-33853 | INTERAGCY | IA-ICS209 | USCAKRN | Kern County Fire Department | NaN | GRAPEVINE | GRAPEVINE | NaN | 2003 | ... | NaN | 1830.0 | F | 34.916944 | -118.918333 | MISSING/NOT SPECIFIED | CA | Kern | 6029.0 | Kern County |
210 rows × 27 columns
### Find out the largest fire in CA, 2020
df.loc[gb['FIRE_SIZE'].idxmax().loc['CA',2020]]
FPA_ID IRW-2020-CAMNF-000730 SOURCE_SYSTEM_TYPE INTERAGCY SOURCE_SYSTEM IA-IRWIN NWCG_REPORTING_UNIT_ID USCAMNF NWCG_REPORTING_UNIT_NAME Mendocino National Forest FIRE_CODE NFP4 FIRE_NAME DOE MTBS_FIRE_NAME AUGUST COMPLEX COMPLEX_NAME AUGUST COMPLEX FIRE_YEAR 2020 DISCOVERY_DATE 8/16/2020 DISCOVERY_DOY 229 DISCOVERY_TIME NaN NWCG_CAUSE_CLASSIFICATION Natural NWCG_GENERAL_CAUSE Natural NWCG_CAUSE_AGE_CATEGORY NaN CONT_DATE 11/11/2020 CONT_DOY 316.0 FIRE_SIZE 589368.0 FIRE_SIZE_CLASS G LATITUDE 39.765255 LONGITUDE -122.672914 OWNER_DESCR USFS STATE CA COUNTY Glenn FIPS_CODE 6021.0 FIPS_NAME Glenn County Name: 400629554, dtype: object
# Plot the number of wildfires every year and associated causes.
df.groupby(['FIRE_YEAR','NWCG_CAUSE_CLASSIFICATION']).FPA_ID.count().unstack('NWCG_CAUSE_CLASSIFICATION').plot(kind = 'bar', stacked = True)
<Axes: xlabel='FIRE_YEAR'>
Exercise 8: Plot the number of wildfires in NJ every year and associated causes. (5 points)¶
3. Assignment: Advanced Pandas with NOAA Hurricane Data¶
3.1 Download the hurricane data from Canvas file. Read in the csv file of NOAA IBTrACS Hurricane Data. (5 points)¶
You are provided with the NOAA IBTrACS dataset file:
Assignment_5_hurricane_ibtracs.since1980.list.v04r00.csv
- Load the dataset into a pandas DataFrame named
df_hurricane. - Only read the first 12 columns from the file.
- Set the first column to be the index column.
- Treat both
-999and blank strings' 'as missing values (NaN).
Hint: refer to the docstring of read_csv function to find out how to set these arguments within the function.
3.2 Get the unique values of the BASIN, SUBBASIN, and NATURE columns (3 points)¶
3.3 Rename the WMO_WIND column to Wind, and WMO_PRES column to Pressure (2 points)¶
3.4 Get the 10 largest rows in the dataset by Wind (3 points)¶
3.5 Group the data on SID and get the 10 largest hurricanes by maximum Wind (5 points)¶
3.6 Plot the count of all datapoints by Basin as a bar chart (5 points)¶
3.7 Plot the count of unique hurricanes by Basin as a bar chart. (5 points)¶